Microfluidics Devices
Large-scale blood processing
Our Microfluidics device employs the principle of hydrodynamic filtration. The advantage is that it is overwhelmingly robust and reproducible, compared with other unlabeled methods such as DLD and Dean Flow.
Traditionally, it has been extremely difficult to flow a blood sample through a Microfluidics device. This is because blood components may clog the inside of the microchannel. We have solved that major problem by adsorbing blood clogging components to beads materials. In addition, the laminated structure of the microfluidic device has made it possible to process more blood in a short time.

High reproducibility and robustness
This microfluidics device also shows excellent performance in recovering tissue-derived cells or a small amount of mouse/rat peripheral blood. Tissue-derived cells obtained through enzymatic treatment contain cell-debris and extra cellular materials. Those components can be removed by the size fractionation using the microfluidics device. Purified cells may be suitable for subsequent cell assays.
This microfluidics device can be sterilized, the recovered cells such like TILs (tumor infiltrating lymphocytes) can be cultured.
<Example 1>
Spike cells mimicking circulating tumor cells (CTC)
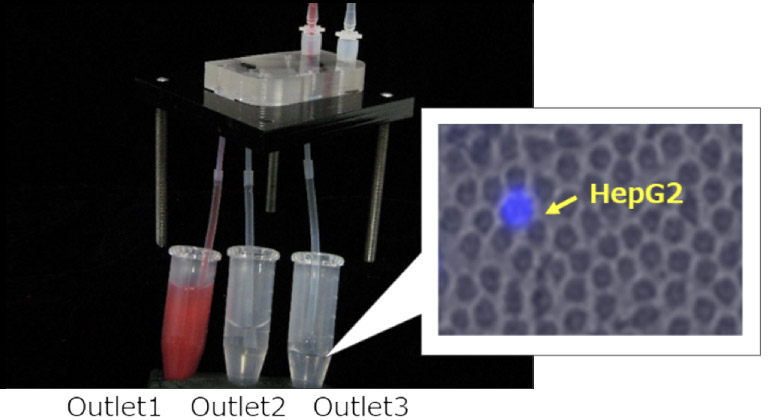
We conducted an experiment to recover HepG2 cells that mimic CTC. HepG2 cells have been stained with Hoechst 33342 in advance, and are aliquoted into small amounts (several cells) by limited dilution. The cell number is actually counted under a fluorescence microscope. In this experiment, 4 cells of HepG2 were spiked into 2 mL of whole blood, and then fractionated by the microfluidics device and then recovered into Outlet3. The running time is just less than 10 minutes.
All the HepG2 cells were recovered in Outlet3, with as nice as 100,000 times enrichment.
Mechanism / performance of Microfluidics device
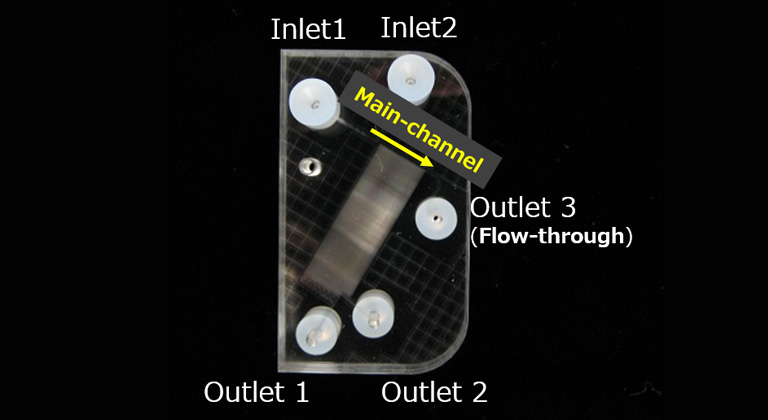
Our microfluidics device has more than 100 side-channels that are crossed with the main-channel. By optimizing the number and width of those side channels, in micrometer ranges, it is possible to separate particles or cells of the desired size. In the example shown, 120 side-channels , two Outlet collection ports (Outet1, Outlet2) and a flow-through (Outlet3) collection port are installed.
When fluorescently labeled beads with sizes of 6 μm, 8 μm, 10 μm, and 15 μm was processed with the microfluidics device, almost all 6 μm beads are collected in Outlet1, almost all 8 and 10 μm beads are in Outlet2, and 84% of 15 μm beads are in Outlet3.

DimShift™
DimShift's Mechanism and Performance
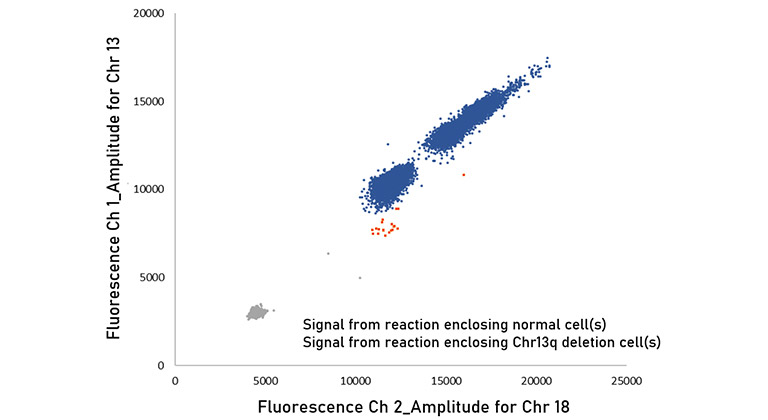
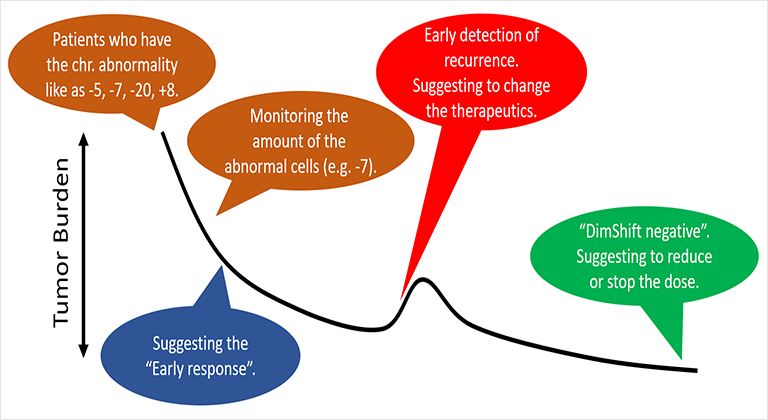
DimShift™ uses single-cell digital PCR technology. Each cell is enclosed in a small compartment (nanoliter size) one by one, and DNA is extracted and amplified from the cells in that compartment, and analysis with our original protocol is performed for each compartment. This allows you to measure the copy number of target gene/locus in each cell. DimShift™ can measure tens of thousands of cells at one time and detect the cells with abnormal copy number. The sensitivity has been investigated as < 0.1%.*1
Figure. The result of DimShift™ (Derived from the mixture of normal cells and 0.1% of cells with Chr13q deletion)
Challenge to Oncology
All tumor cells forming cancer have abnormality in their DNA.
DNA abnormalities vary from single nucleotide mutations, deletions, amplifications and aneuploidies, etc
Among them, we focus on deletions and amplifications (copy number variations: CNV). Deletions and amplifications occur frequently in major solid tumors and hematological malignancies (Example: Chr13q deletion occurs in around 50% of MM and in around 80% of CLL, and Chr18q deletion has been reported in about 50% of cases of colorectal cancer)reference 1, 2
Although “Cytogenetic analysis” such as FISH and G-banding or “Molecular analysis” such as PCR and NGS can be used to detect CNV, sensitivity of these has been limited because the number of cells to be analyzed is limited in FISH and G-banding and the DNA from tumor is to be diluted with normal DNA in PCR or NGS*2
We are solving that problem by increasing the sensitivity around 100 times with “DimShift™.
This makes it possible to detect minimal residual disease (MRD) that exists in the body after treatment, which may be useful for continuing, changing, or discontinuing treatment*3
Reference1 The Japanese Society of Internal Medicine, 100, 7, July 10th, 201
Reference2 BMC Cancer 2007, 7:226
*1 Percentage of abnormal cells in analyzed cells.
*2 PCR and NGS analyze the average value of the number of copies of the target of the cell population for the DNA extracted from the analysis cell population. Therefore, when the proportion of normal cells in the population increases, the measured value (mean value) Is no different from normal, and it becomes indistinguishable whether or not abnormal cells were present.
*3DimShift™ related products are Research Use Only and cannot be used for clinical diagnosis.
Contribution to cell therapy and biotechnology-based therapeutics
DimShift™ may be useful for the production, quality control and post-administration evaluation of cells for gene therapy or antibody production. In recent years, contributions of cell therapy products such as CAR-T therapy to cancer treatment are accelerating. In order to provide safer and more effective treatments, we are working on the development of the single cell copy number counting system for CAR gene by DimShift™. Furthermore, we are trying to contribute the quality control of cell line (e.g. CHO cell line) for the better production of biotechnology-based therapeutics such as antibody drugs.

DimScat™
Our approach (Prenatal testing)
The definitive diagnosis for chromosomal abnormalities in fetus is performed by amniocentesis test*1 or Chorionic villus sampling (CVS) test*2. Traditionally, amniocentesis and CVS have been performed by the G-banding*3, which is performed by culturing cells brought from amniotic fluid or chorionic villus. Recently, technologies such as microarray*4 and the QF-PCR*5 have been used to investigate chromosomal abnormalities more easily and quickly.
However, these methods still have problems in terms of measurement time, coverage of chromosomal aberrations, and testing cost.
We are solving some of these challenges by DimScat.
This makes it possible to measure the aneuploidies of chr13, chr18, chr21 and sex chromosome as well as various micro deletions including 22q11 deletion with shorter turnaround time (4 hours). Since it can be tested by one or two tubes, it also has the effect of reducing costs.
“The basic panel of 16 multiplex for chr13, 18, 21, 22q11, X, Y” and “The extended panel of 9 multiplex for chr 5p, 15q11-13, 18” are our line-ups.
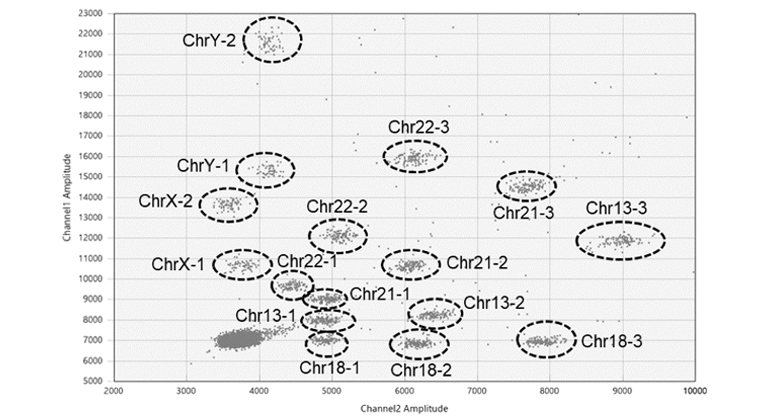
Figure. Basic panel. The letters next to the cluster are the target chromosome locus ID (multiple locus are measured for each chromosome to improve quality).
*1 A method for detecting abnormalities of chromosomes in fetal cells in amniotic fluid corrected from the endometrium of the mother. Amniotic fluid contains a large amount of fetal cell, and the accuracy of diagnosis is high and it is considered to be a definitive test.
*2 A test collects and cultures villi, which are part of the placenta in early pregnancy, and detects the number of chromosomes. It is a definitive test.
*3 Cultivate the cells in the sample and measure the number of chromosomes or chromosomal regions. Deletions or amplifications of the entire chromosome or large regions can be detected, but micro deletions or amplifications cannot be detected due to the resolution.
*4 A microarray is a laboratory tool used to detect the copy of thousands of genes at the same time using the chip in which DNA molecules are attached. It can also inspect more comprehensively and detect micro deletions or amplifications. In general, the cost increase accordingly.
*5 This is a method to detest the number of chromosomes by amplifying and quantifying specific repeating sequences (STR) in DNA.
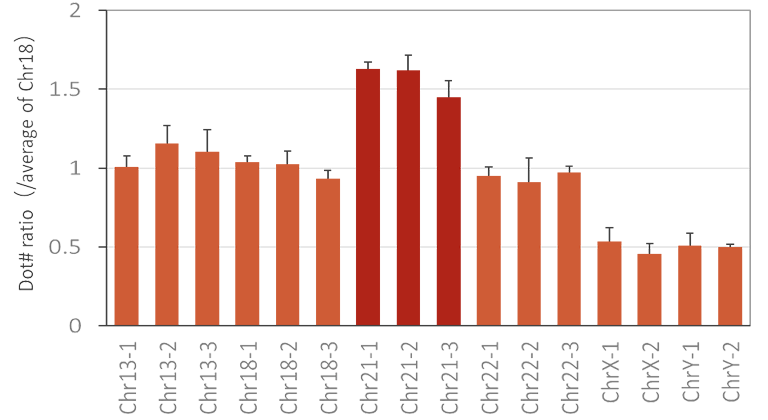
Figure. Measurement results of DNA derived from trisomy 21 cells (Detorit532).